Comparative Protein Expression Analysis for Precision Diagnosis of Multiple Sclerosis
Gene coexpression with transcriptome analysis, a measure of what genes are actually expressed in an individual, has been used to distinguish patients with relapsing-remitting multiple sclerosis (RRMS) from healthy control subjects. This analysis was also able to distinguish the pattern of distinct immune cell profiles in patients with MS vs healthy controls.
Compared with healthy control subjects, patients with RRMS had significantly higher coexpression of T-cell networks, lower levels of B-cell network, monocytes, interferon-induced cells, and other immune cell types (Table).
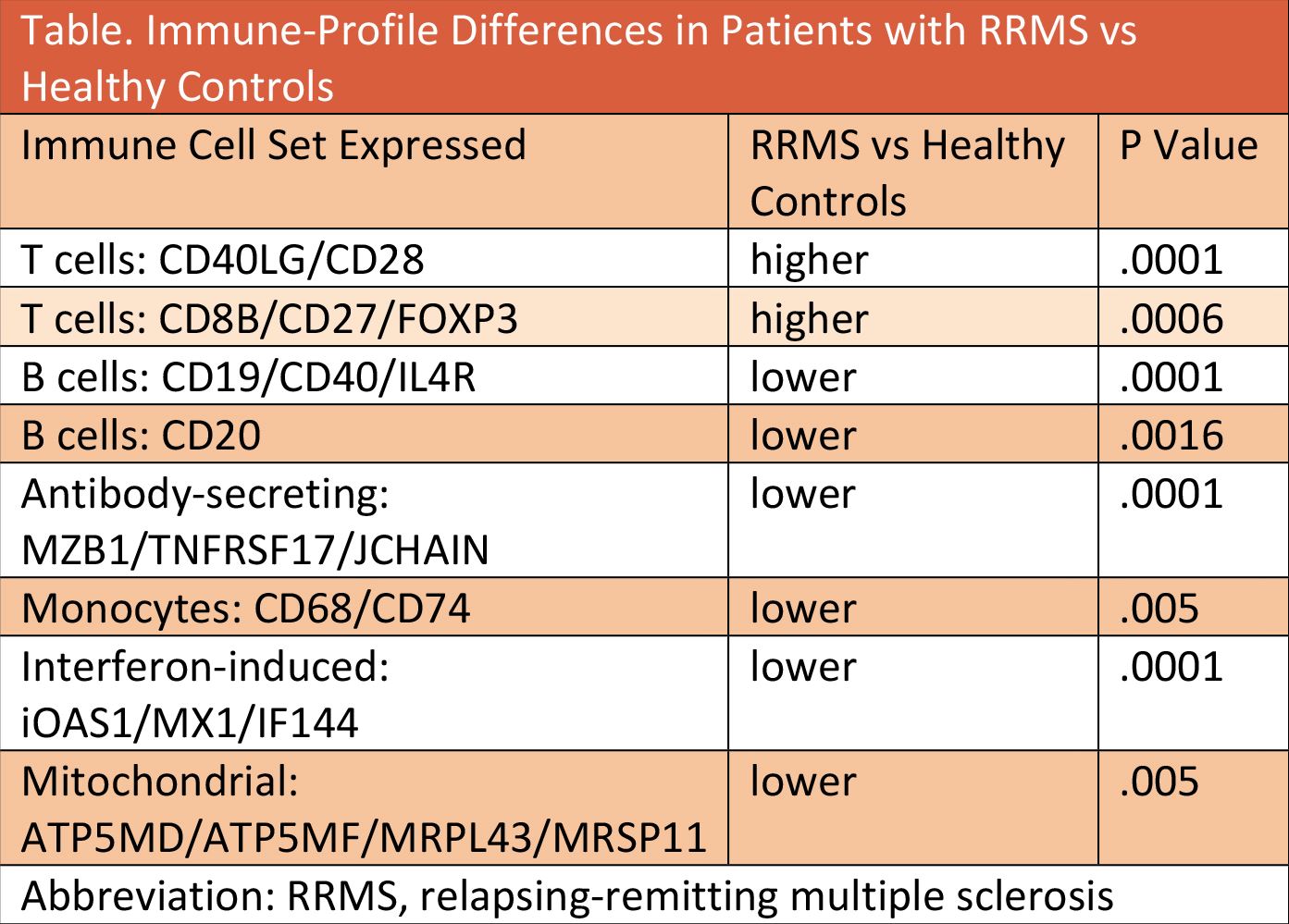
These data come from the CARE-MS trials (NCT00530348and NCT00548405) studying alemtuzumab vs interferons. Before beginning treatment in any arm of the trial, peripheral blood mononuclear cells (PBMCs) were collected from patients with RRMS (n = 216) and from healthy control subjects in the Genotype-Tissue Expression project (n = 392). Transcriptome analysis with RNA sequencing was used to identify the immune profiles of all subjects. These data were presented at the American Conference on Treatment and Research in MMS (ACTRIMS) conference in Dallas, TX February 28-March 1.
